| pre | post |
|---|---|
| 5260 | 3910 |
| 5470 | 4220 |
| 5640 | 3885 |
| 6180 | 5160 |
| 6390 | 5645 |
| 6515 | 4680 |
Elements of Statistical Inference, Part II
September 3, 2025
Comparing two population parameters
Two-sample data can be paired or unpaired (independent).
Paired measurements for each “participant” or study unit
- each unit can be matched to another unit in the data
- e.g., “enriched” vs. “normal” environments for pairs of rats, with pairs taken from the same litter
Two independent sets of measurements
- observations cannot be matched on a one-to-one basis
- e.g., rats from several distinct litters observed in either “enriched” or “normal” environments
Nature of the data dictates testing procedure: two-sample test for paired data or two-sample test for independent group data.
Example: Menstruation’s effect on energy intake
Does the dietary (energy) intake of women differ pre- versus post-menstruation (across 10 days)? (Table 9.3 of Altman 1990)
A study was conducted to assess the effect of menstruation on energy intake.
- Investigators recorded energy intake for each of the women (in kJ)
The paired
The energy intake measurements are paired within a given study unit (each of the women)—use this structure to advantage, i.e., exploit homogeneity.
For each woman
Define the measurement difference:
Base inference on
The paired
Let
The null and alternative hypotheses are
- i.e., no effect of menstruation on subsequent energy intake
- i.e., menstruation does have an effect on energy intake
The paired
The test statistic for the paired
A paired
Letting R do the work
Paired
Paired t-test
data: intake$pre and intake$post
t = 11.941, df = 10, p-value = 3.059e-07
alternative hypothesis: true mean difference is not equal to 0
95 percent confidence interval:
1074.072 1566.838
sample estimates:
mean difference
1320.455 Differences
# one-sample syntax
t.test(
intake[, pre - post],
mu = 0, paired = FALSE,
alternative = "two.sided"
)
One Sample t-test
data: intake[, pre - post]
t = 11.941, df = 10, p-value = 3.059e-07
alternative hypothesis: true mean is not equal to 0
95 percent confidence interval:
1074.072 1566.838
sample estimates:
mean of x
1320.455 Independent
# two-sample syntax
t.test(
intake$pre, intake$post,
mu = 0, paired = FALSE,
alternative = "two.sided"
)
Welch Two Sample t-test
data: intake$pre and intake$post
t = 2.6242, df = 19.92, p-value = 0.01629
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
270.5633 2370.3458
sample estimates:
mean of x mean of y
6753.636 5433.182 Assumptions for paired
Like the one-sample version, paired
- Differences arise as iid samples from
- …assumed to have zero mean (under
- …assumed to have unknown variance (i.e.,
- …assumed to have zero mean (under
- Additionally requirements include independence of pairs and non-interference (that is, one pair cannot affect another)
- For example, difference of pre- and post-menstruation energy intake assumed to be normally distributed
FAMuSS: Comparing arm strength by sex
Question: Does change in non-dominant arm strength (ndrm.ch) after resistance training differ between men and women?
| sex | age | height | weight | actn3.r577x | ndrm.ch |
|---|---|---|---|---|---|
| Female | 27 | 65.0 | 199 | CC | 40 |
| Male | 36 | 71.7 | 189 | CT | 25 |
| Female | 24 | 65.0 | 134 | CT | 40 |
| Female | 40 | 68.0 | 171 | CT | 125 |
| Female | 32 | 61.0 | 118 | CC | 40 |
| Female | 24 | 62.2 | 120 | CT | 75 |
famuss dataset from Vu and Harrington (2020)
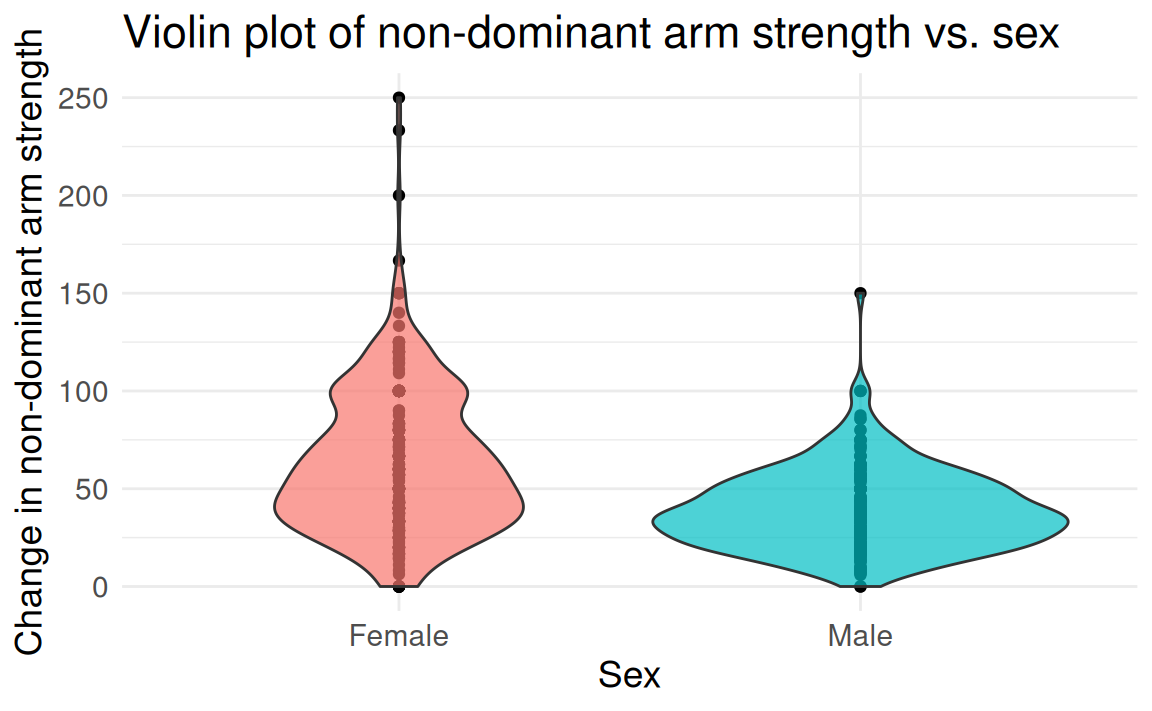
The two-group (independent)
Framing the question — the null and alternative hypotheses are
- Equivalently, let
- Equivalently, let
Hypotheses may generically be written in terms of
- The parameter of interest is
- The point estimate is
The two-group (independent)
The test statistic for the two-group (independent)
Two-group (independent)
The two-sample
A conservative approximation is
The Welch-Satterthwaite approximation1 (used by R):
Alternative is to use Student’s version
Confidence intervals for the two-group (independent) difference-in-means
A
Letting R do the work
Welch Two Sample t-test
data: famuss$ndrm.ch[female] and famuss$ndrm.ch[male]
t = 10.073, df = 574.01, p-value < 2.2e-16
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
19.07240 28.31175
sample estimates:
mean of x mean of y
62.92720 39.23512 uses
Two Sample t-test
data: famuss$ndrm.ch[female] and famuss$ndrm.ch[male]
t = 9.1425, df = 593, p-value < 2.2e-16
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
18.60259 28.78155
sample estimates:
mean of x mean of y
62.92720 39.23512
uses1
Assumptions for two-group (independent)
- Observations arise as iid samples from
- …assumed to have same population mean (under
- …assumed to have same unknown variance2 (i.e.,
- …assumed to have same population mean (under
- Additional requirements include independence of observations and non-interference (one observation cannot affect another)
- For example, change in non-dominant arm strength for men and women is assumed to follow
Permutation-based hypothesis testing
Permutation testing1 is a nonparametric framework for inference
- aim is to limit assumptions about the underlying populations, instead using structure that can be induced by design
- method: construct the null distribution of a test statistic via (artificial) randomization, as if assignment2 was controlled
Permutation/randomization inference evaluates evidence against a different null hypothesis (
Assumptions for permutational
- Unlike the paired or two-group
- …there is no need to assume a specific distributional form for how the data—observations or paired differences—arise
- …non-interference issues (units cannot affect each other) are resolved by the construction of the null hypothesis
- So…what do we need then?
- Groups (or differences) are exchangeable (under
- Randomization (of assignment) is performed fairly
- Groups (or differences) are exchangeable (under
The permutational
Exact Two-Sample Fisher-Pitman Permutation Test
data: ndrm.ch by sex (Female, Male)
Z = 8.5664, p-value < 2.2e-16
alternative hypothesis: true mu is not equal to 0Exact: How many ways to choose
Approximative Two-Sample Fisher-Pitman Permutation Test
data: ndrm.ch by sex (Female, Male)
Z = 8.5664, p-value < 1e-04
alternative hypothesis: true mu is not equal to 0Approximate: What about 10000? Is that enough?
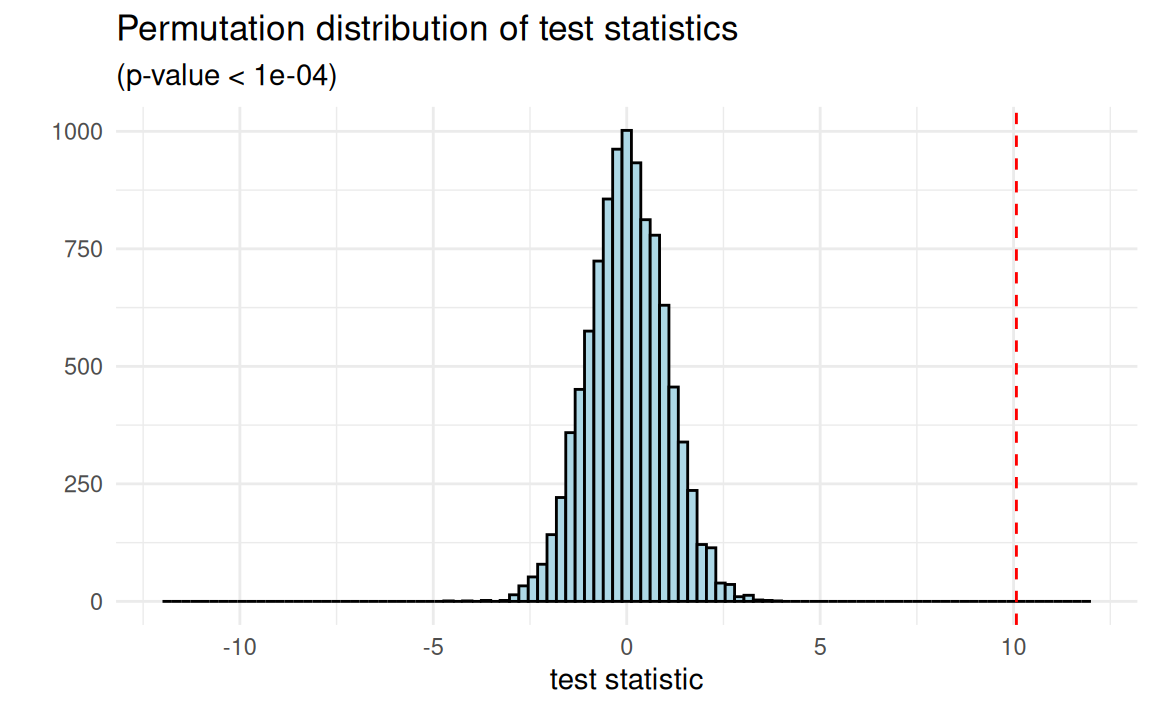
Null hypotheses: The weak and the sharp
Data on
Weak null: averages
Is there no effect on average?
Sharp (strong) null: individuals
Is there no effect for everyone?
The sharp null implies the weak null—no effect at all, no effect on average too.
FAMuSS: Comparing arm strength by genotype
Going beyond two-group comparisons
Is change in non-dominant arm strength after resistance training associated with genotype?
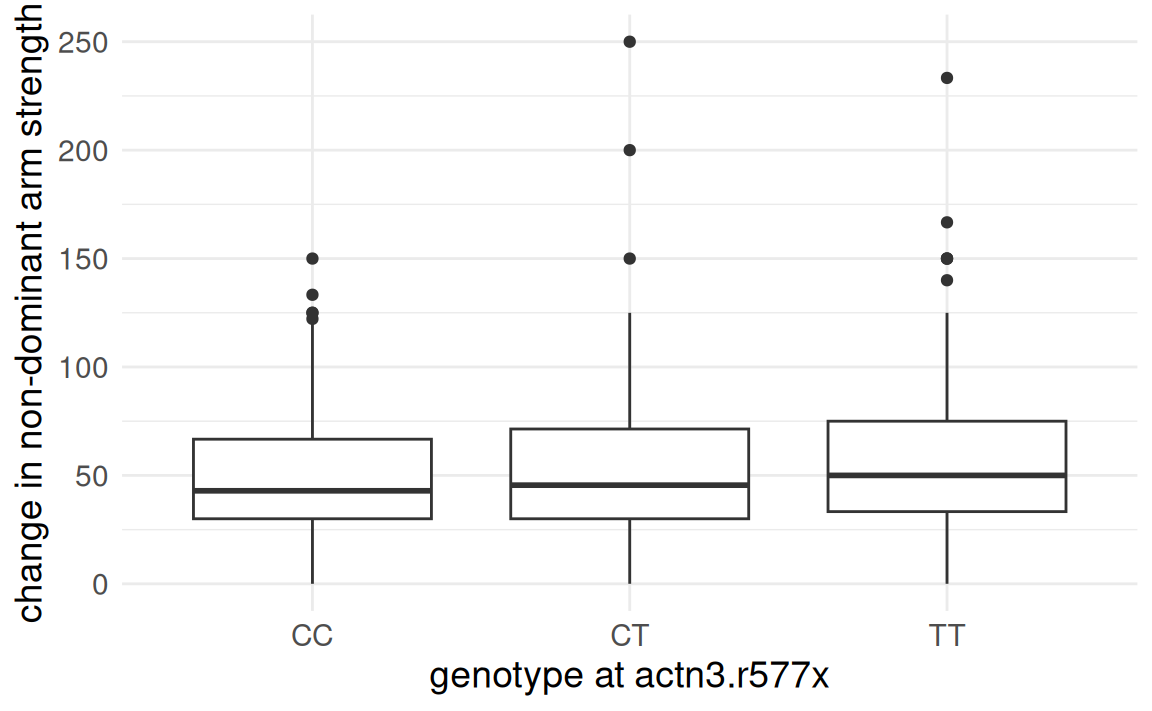
Analysis of Variance (ANOVA)
Suppose we are interested in comparing means across more than two groups. Why not conduct several two-sample
- If there are
- Conducting multiple tests on the same data increases the overall rate of Type I error, necessitating a multiplicity correction.
ANOVA: Assesses equality of means across many groups:
ANOVA: Are the groups all the same?
Under
- Think of all observations as belonging to a single, large group.
- Variability between group means
ANOVA exploits differences in means from an overall mean and within-group variation to evaluate equality of a few group means.
ANOVA: Are the groups all the same?
Is the variability in the sample means large enough that it seems unlikely to be due to chance alone?
Compare two quantities:
- Variability between groups (
- Variability within groups (
ANOVA: The
The
ANOVA measures discrepancies via the F-statistic, which follows an
- The
Assumptions for ANOVA
Just like the
- Observations are independent within and across groups
- Data within each group arise from a normal distribution
- Variability across the groups is about equal
When these assumptions are violated, hard to know whether
As with the
Pairwise comparisons
If the
Pairwise comparisons may use the two-group (independent)
- To maintain overall Type I error rate at
- The Bonferroni correction1 is one method for adjusting
- Note: The Bonferroni correction is conservative (i.e., stringent), and assumes all tests are independent.
Letting R do the work
Df Sum Sq Mean Sq F value Pr(>F)
famuss$actn3.r577x 2 7043 3522 3.231 0.0402 *
Residuals 592 645293 1090
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Conclusion:
But which group(s) is (are) the source of the difference? 🧐
Controlling Type I error rate
If the
- Each test should be conducted at the
- These
- We will use
pairwise.t.test()to perform these post hoc two-sample t-tests.
Controlling Type I error rate
Pairwise comparisons using two-sample CC to CT, CC to TT, CT to TT) can now be done if the Type I error rate is controlled.
- Apply the Bonferroni correction.
- In this setting,
Examine each of the three two-sample
Letting R do the work
Only CC versus TT resulted in a
- Mean strength change in non-dominant arm for individuals with genotype
CTis not distinguishable from strength change for those withCCandTT. - However, evidence at
CCandTTare different.
Type I error rate for a single test
Hypothesis testing was intended for controlled experiments or for studies with only a few comparisons (e.g., ANOVA).
Type I errors (rejecting
- Type I error rate is controlled by rejecting only when the
- With a single two-group comparison at
And what about many tests?
Multiple testing–compounding error
What happens to Type I error when making several comparisons? When conducting many tests, many chances to make a mistake.
- The significance level (
- Experiment-wise error rate: the chance of at least one test incorrectly rejecting
- Controlling the experiment-wise error1 rate is just one approach for controlling the Type I error.
Probability of experiment-wise error
A scientist is using two
- Control
Example of compounding of Type I error
Probability of making at least one error is equal to the complement of the event that a Type I error is not made with either test.
Probability of experiment-wise error…
10 tests…
25 tests…
100 tests…
With 100 independent tests: 99.4% chance an investigator will make at least one Type I error!
Advanced melanoma
Advanced melanoma is an aggressive form of skin cancer that until recently was almost uniformly fatal.
Research is being conducted on therapies that might be able to trigger immune responses to the cancer that then cause the melanoma to stop progressing or disappear entirely.
In a study where 52 patients were treated concurrently with 2 new therapies, nivolumab and ipilimumab, 21 had immune responses.1
Advanced melanoma
Some research questions that can be addressed with inference…
- What is the population probability of immune response following concurrent therapy with nivolumab and ipilimumab?
- What is a 95% confidence interval for the population probability of immune response following concurrent therapy with nivolumab and ipilimumab?
- In prior studies, the proportion of patients responding to one of these agents was 30% or less. Do these data suggest a better (>30%) probability of response to the concurrent therapy?
Inference for binomial proportions
In this study of melanoma, experiencing an immune response to the concurrent therapy is a binary event (i.e., binomial data).
Suppose
- Goal: Inference about population parameter
Assumptions for using the normal distribution
The sampling distribution of
- The sample observations are independent, and
- At least 10 successes and 10 failures are expected in the sample:
Under these conditions,
Inference with the normal approximation
For CIs, use
In hypothesis testing:
Test statistic
1-sample proportions test with continuity correction
data: 21 out of 52, null probability 0.3
X-squared = 2.1987, df = 1, p-value = 0.06906
alternative hypothesis: true p is greater than 0.3
95 percent confidence interval:
0.2906582 1.0000000
sample estimates:
p
0.4038462 Exact inference for binomial data
An exact procedure does not reply upon a limit approximation.1
The Clopper-Pearson CI is an exact method for binomial CIs:
Note: While exactness is by construction, the CIs can be conservative (i.e., too large) relative to those from an approximation.
[1] 0.07167176
Exact binomial test
data: 21 and 52
number of successes = 21, number of trials = 52, p-value = 0.07167
alternative hypothesis: true probability of success is greater than 0.3
95 percent confidence interval:
0.2889045 1.0000000
sample estimates:
probability of success
0.4038462 Inference for difference of two proportions
The normal approximation can be applied to
- The samples are independent, the observations in each sample are independent, and
- At least 10 successes and 10 failures are expected in each sample.
The standard error of the difference in sample proportions is
Treating HIV
In resource-limited settings, single-dose nevirapine is given to HIV
- Exposure of the infant to nevirapine (NVP) may foster growth of resistant strains of the virus in the child.
- If the child is HIV
Here, the possible outcomes are virologic failure (virus becomes resistant) versus stable disease (virus growth is prevented).
Treating HIV
The results of a study comparing NVP vs. LPV in treatment of HIV-infected infants.1 Children were randomized to receive NVP or LPV.
| NVP | LPV | Total | |
|---|---|---|---|
| Virologic Failure | 60 | 27 | 87 |
| Stable Disease | 87 | 113 | 200 |
| Total | 147 | 140 | 287 |
Is there evidence of a difference in NVP vs. LPV? How to see this?2
Formulating hypotheses in a two-way table
Do data support the claim of a differential outcome by treatment?
If there is no difference in outcome by treatment, then knowing treatment provides no information about outcome, i.e., treatment assignment and outcome are independent (i.e., not associated).
Question: What would we expect if no association (i.e., under
The
Idea: How do the observed cell counts differ from those expected (under
The Pearson
- Large test statistic: Stronger evidence against the null hypothesis of independence.
- Small test statistic: Weaker evidence (or lack of) against the null hypothesis of independence.
Assumptions for the
- Independence: Each case contributing a count to the
- Sample size: Each expected cell count must be greater than or equal to 10.1 (Without enough data, what are you comparing?)
Under these assumptions, the Pearson
The
The
The
Applying the
If treatment has no effect on outcome, what would we expect?
Under the null hypothesis of independence,
Same logic applies for other cells…repeat three times and sum to get the
The R
The data from Violari et al. (2012):
What would we expect to see under independence? (Table of
NVP LPV
Virologic Failure 44.56 42.44
Stable Disease 102.44 97.56By comparing cells of these two tables (of
R’s chisq.test() does all of this:
Pearson's Chi-squared test with Yates' continuity correction
data: hiv_table
X-squared = 14.733, df = 1, p-value = 0.0001238Conclusion: The
Fisher’s exact test
R.A. Fisher proposed an exact test for contingency tables by exploiting the counts and margins, with the “reasoned basis” for the test being randomization and exchangeability (Fisher 1936).
| NVP | LPV | Total | |
|---|---|---|---|
| Virologic Failure | a | b | a+b |
| Stable Disease | c | d | c+d |
| Total | a+c | b+d | n |
Here,
R’s fisher.test() does this:
Fisher's Exact Test for Count Data
data: hiv_table
p-value = 0.0001037
alternative hypothesis: true odds ratio is not equal to 1
95 percent confidence interval:
1.643280 5.127807
sample estimates:
odds ratio
2.875513 Conclusion: Evidence for independence of NPV, virologic failure lacking (
Lowering stakes: The lady tasting tea
“Dr. Muriel Bristol, a colleague of Fisher’s, claimed that when drinking tea she could distinguish whether milk or tea was added to the cup first (she preferred milk first). To test her claim, Fisher asked her to taste eight cups of tea, four of which had milk added first and four of which had tea added first.” Agresti (2012)
| Tea first (real) | Milk first (real) | Total | |
|---|---|---|---|
| Tea first (thinks) | 2 | 2 | 4 |
| Milk first (thinks) | 2 | 2 | 4 |
| Total | 4 | 4 | 8 |
What’s the probability of getting all four right?
| Tea first (real) | Milk first (real) | Total | |
|---|---|---|---|
| Tea first (thinks) | 4 | 0 | 4 |
| Milk first (thinks) | 0 | 4 | 4 |
| Total | 4 | 4 | 8 |
Fisher's Exact Test for Count Data
data: ladys_tea
p-value = 0.02857
alternative hypothesis: true odds ratio is not equal to 1
95 percent confidence interval:
1.339059 Inf
sample estimates:
odds ratio
Inf The relative risk in a
Relative risk (RR) measures the risk of an event occurring in one group relative to risk of same event occurring in another group.
The risk of virologic failure among the NVP group is
The risk of virologic failure among the LPV group is
RR of virologic failure for NVP vs. LPV is
The odds ratio in a
Odds ratio (OR) measures the odds of an event occurring in one group relative to the odds of the event occurring in another group.
The odds of virologic failure among the NVP group is
The odds of virologic failure among the LPV group is
OR of virologic failure for NVP vs. LPV is
Relative risk versus odds ratio
The relative risk cannot be used in studies that use outcome-dependent sampling, such as a case-control study:
- Suppose in the HIV study, researchers had identified 100 HIV-positive infants who had experienced virologic failure (cases) and 100 who had stable disease (controls), then recorded the number in each group who had been treated with NVP or LPV.
- With this design, the sample proportion of infants with virologic failure no longer estimates the population proportion (it is biased by design).
- Similarly, the sample proportion of infants with virologic failure in a treatment group no longer estimates the proportion of infants who would experience virologic failure in a hypothetical population treated with that drug.
- The odds ratio remains valid even when it is not possible to estimate incidence of an outcome from sample data.
References

HST 190: Introduction to Biostatistics